
 |
Browse Motifs | Help/About | View Cart | |
| Selected Version: 1.02 - CHANGE | ||||
This page helps to showcase what the site has to offer. From the home page, you can see two menus: The top menu and the side menu.

At any point you can click the "YeTFaSCo" logo at the left side of the top menu to get back to the home page.

Say you want to view the motifs in the database and how they are scored. To do this you can either click "Browse Motifs" in the top menu, or "Browse Tables">"Motifs" on the side menu.
Now you should see the first page of the Motifs table that should look something like this:
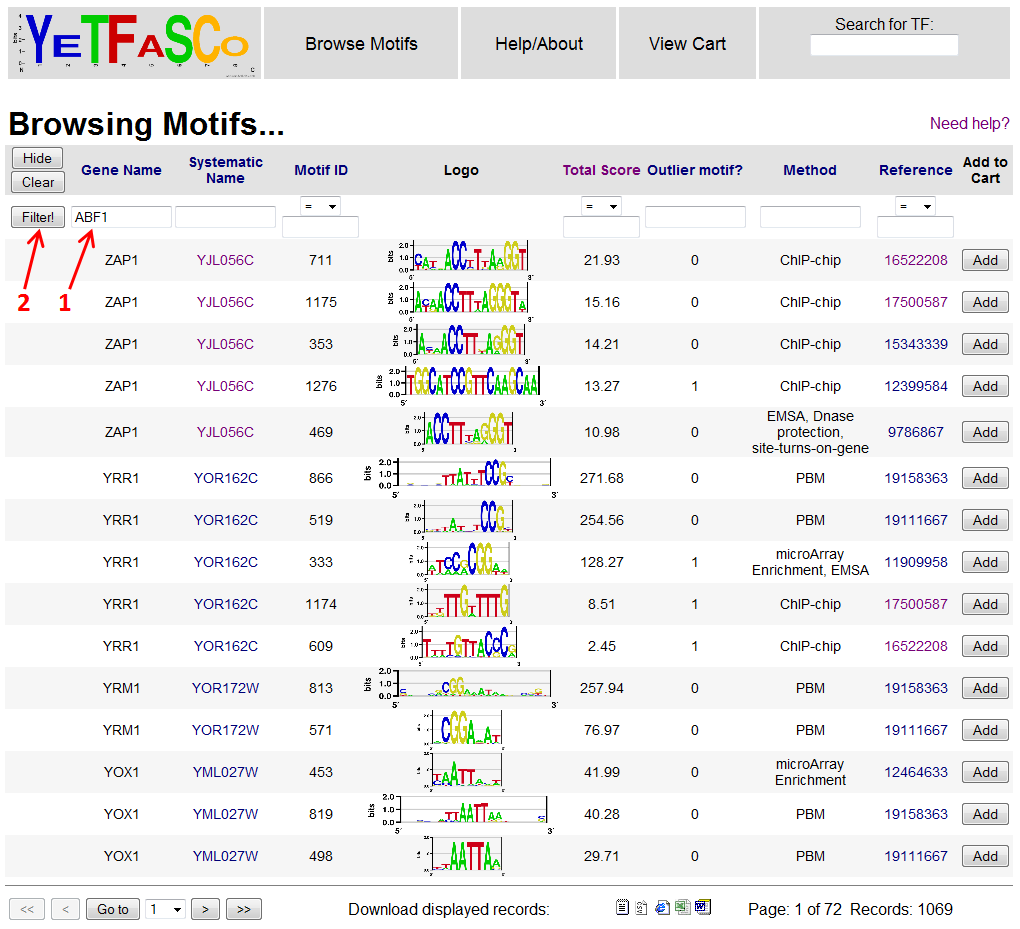
Say you're interested in a particular transcription factor, say ABF1. You can narrow down the motifs displayed by adding the gene name into the filter field (1), and then pressing enter or clicking the "Filter!" button (2). Now you should only be viewing the records for your gene of interest.
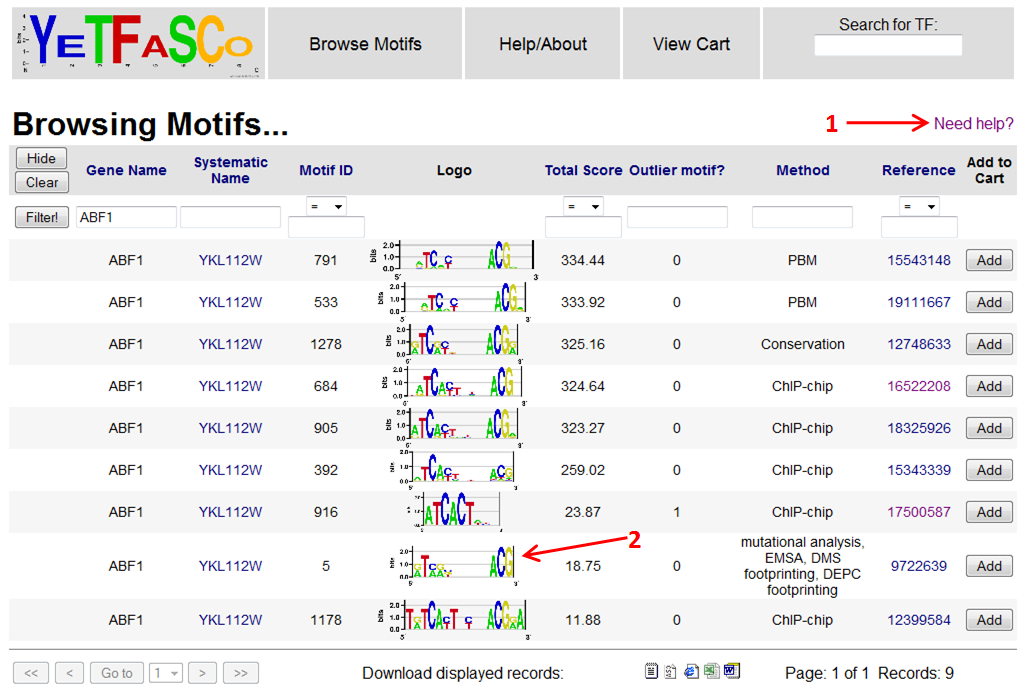
To get a further explanation of what each of the columns mean, you can click the "Need Help?" link in the upper right (1). Once you've read about the column names, you can click the "Back" button on your browser to return to the search results. Maybe you want to see why one motif got a particularly good or bad score. To see the details of how the motif was scored, you can click the motif logo link (2).
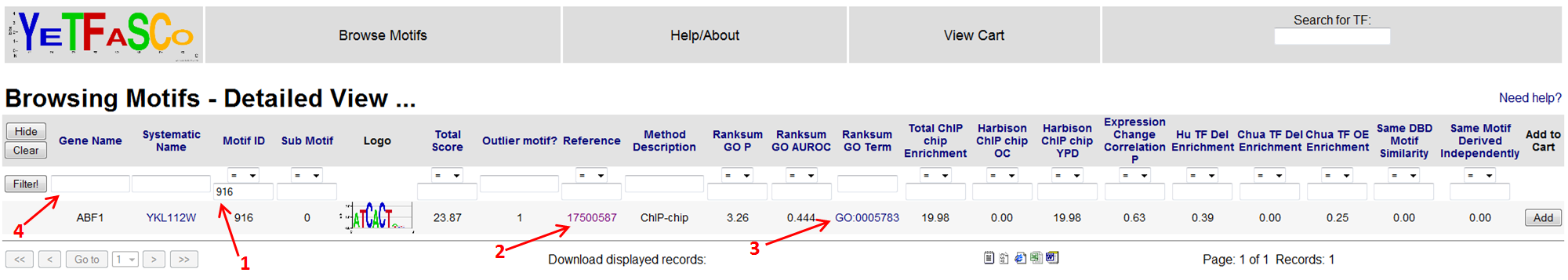
You can see in the detailed view the filter that is being applied (1). In this case it is the unique identifier for the motif. If you want to see details of the study which derived this motif, you can click on the reference link (2). This will bring you to the References table.
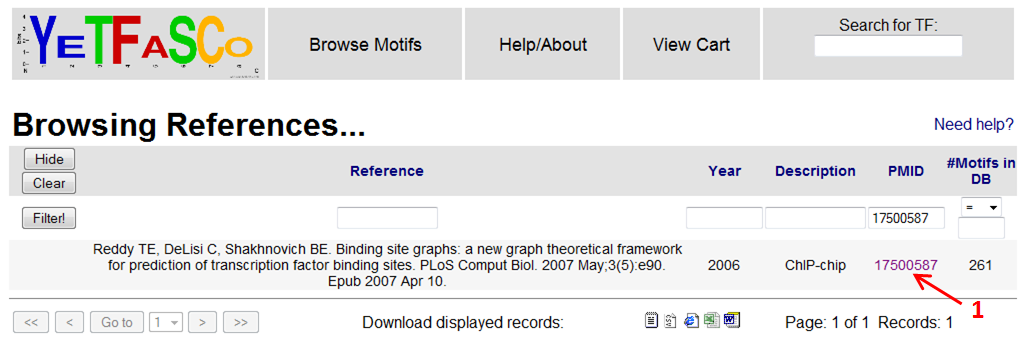
From here, you can click on the PMID to get to PubMed where you can download the paper (1). You can then press "Back" to return to the detailed motif view.

From detailed motif view, you can also click the GO term link (3) which will bring you to the GO page describing that GO term. You can also change the filter here, as before, to view all the other motifs for your gene of interest by changing the contents of the filter bar (4). In this case, you would also have to clear the Motif ID filter currently being applied (1).
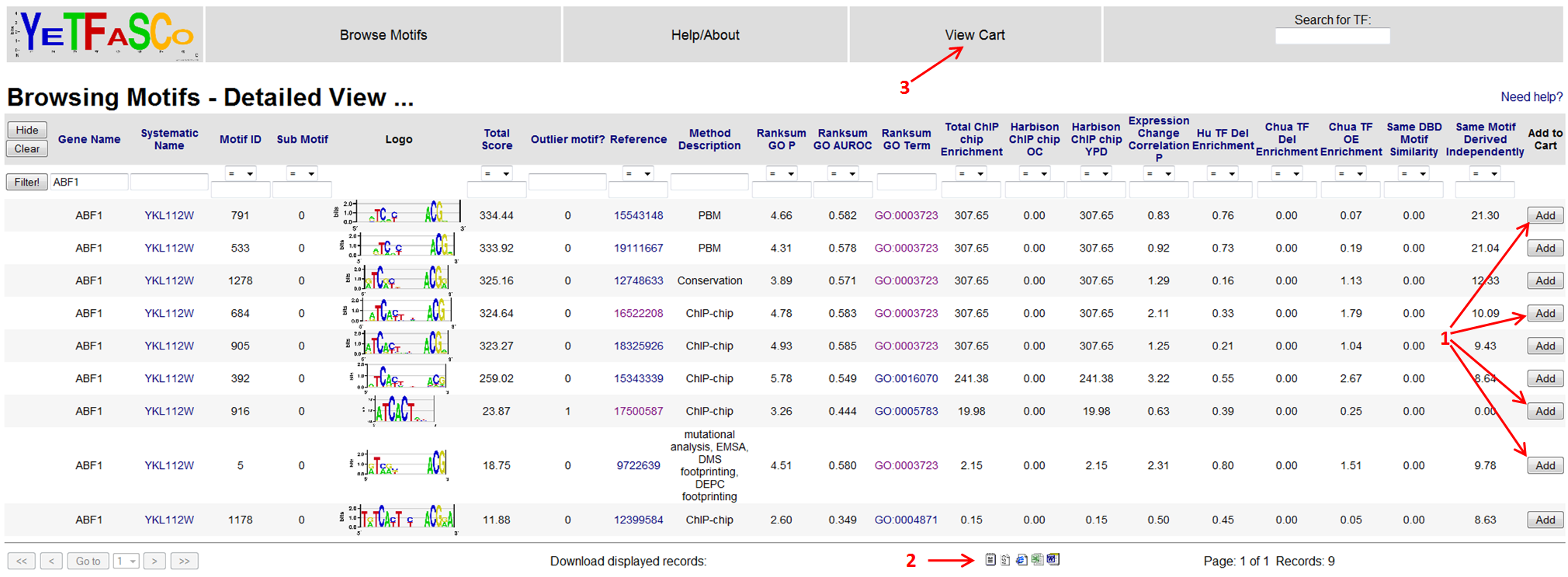
Say you want to hang on to some of these results for the moment. You can do this by adding them to your cart. This is done by clicking the "Add" buttons for each motif (1). Additionally, for all tables, you can download the data in various formats by clicking the icons at the bottom (2). To see the contents of your cart, you can click the "View Cart" link at the top of the page (3).
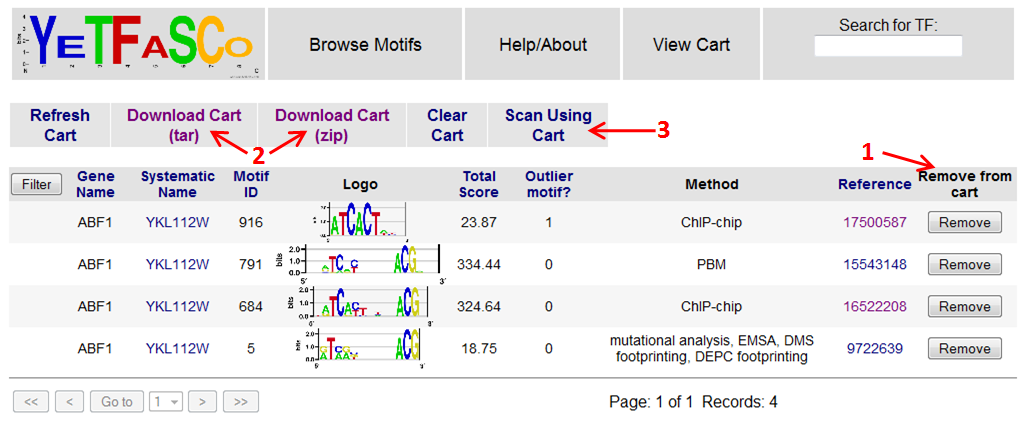
From the cart, you can remove motifs from your cart (1), download the position frequency matrices for the motifs in your cart (2), and scan sequences using your cart (3). The "Scan Using Cart" link takes you to the sequence scanning page.
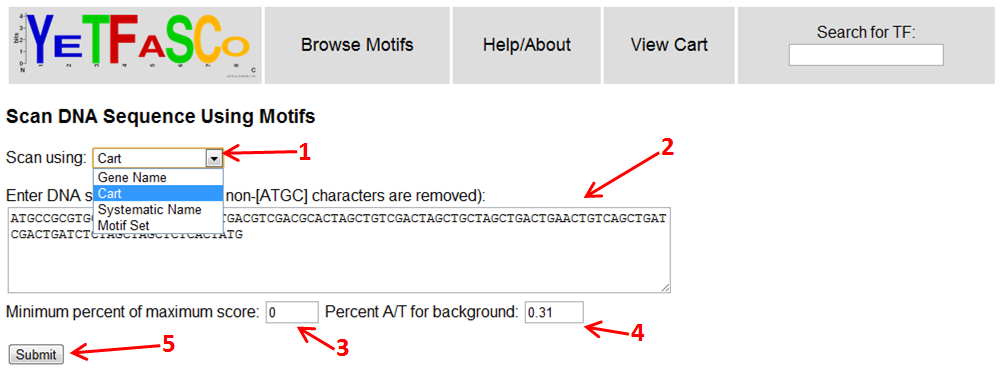
You can choose to scan DNA sequences with your cart, by gene or systematic name (for one TF or one motif), or by one of the provided non-redundant motif sets (1). Once you select the motifs to use for scanning, you can then enter your DNA sequence (2) and press the "Submit" button (5) to scan the DNA. There are also options to threshold the hits by the percent of the maximum score (3) and to alter the background base content (4).
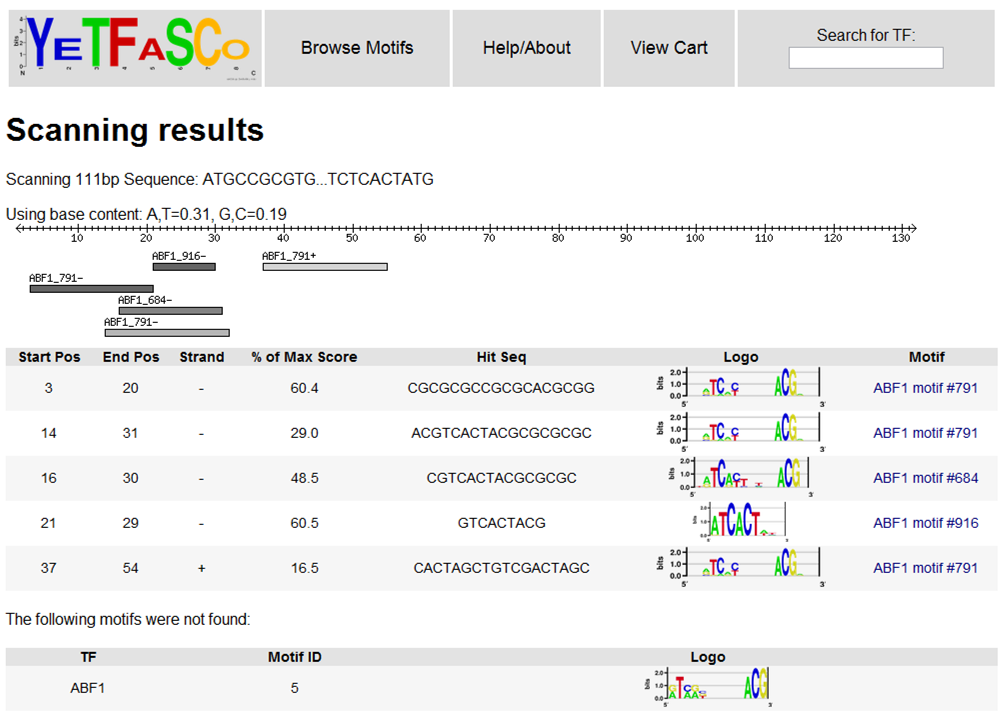
Here, you can see the locations of the predicted binding sites, the sequence that matched the motif, and the percent of the maximum possible score that sequence achieved.
Say you found a motif upstream of some of your genes of interest and want to see if a TF is known to bind that sequence. You can scan the database for similar motifs to your query motif using the "Find Similar Motifs" tool found under tools on the side menu of the homepage.
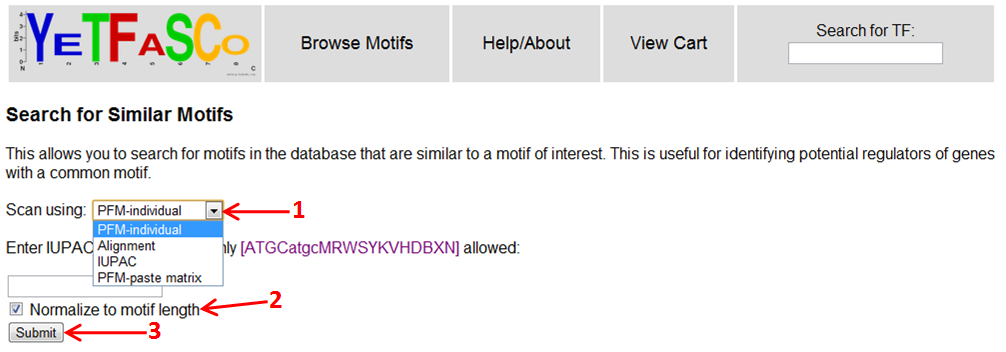
There are three different ways you can enter your query motif: by PFM (either by filling out the individual frequencies, or by pasting in a PFM), by sequence alignment, and by IUPAC motif (1). You can also choose whether or not to normalize to the length of the motif (2). This has the effect of giving you a per-base distance measure in contrast to a total distance measure where longer PFMs tend to have a greater total distance. Once you enter in your motif in whatever form you like, you can click Submit to see your results (3).
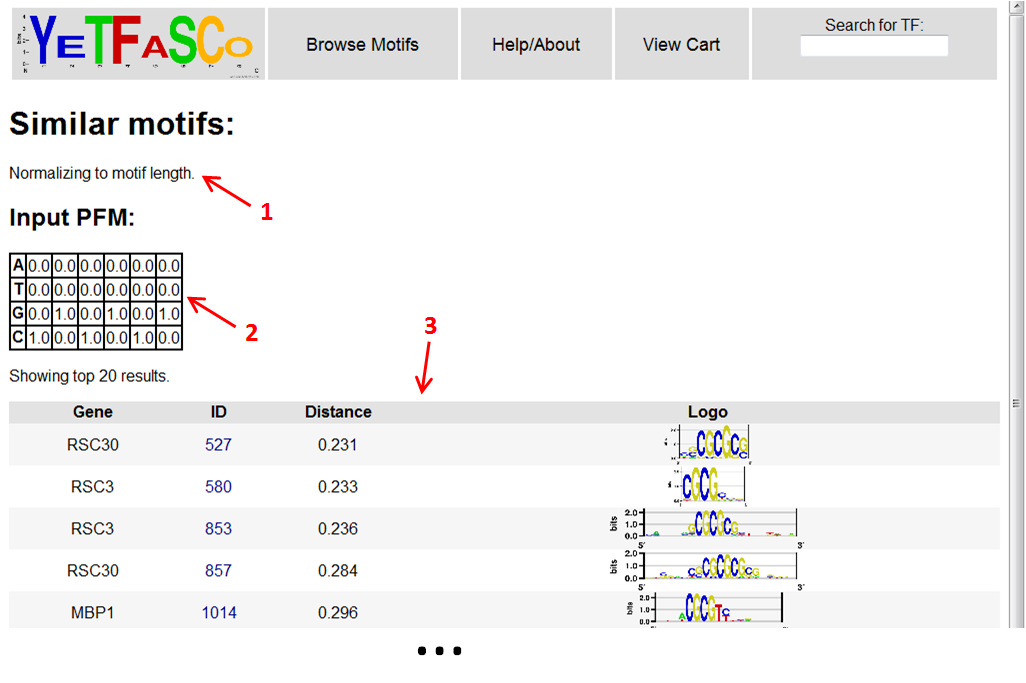
Once the results are ready, it will display whether or not you chose to normalize to motif length (1), what your motif looks like as a PFM (2), and the top 20 closest motifs in the database to yours (3). From here, you can click on the motif IDs to get to their entry in the motif table.
In addition, the website also has precomputed genome-wide binding site predictions available via genome-browser. This feature is available on the main page side bar as "Binding Site Genome Browser".
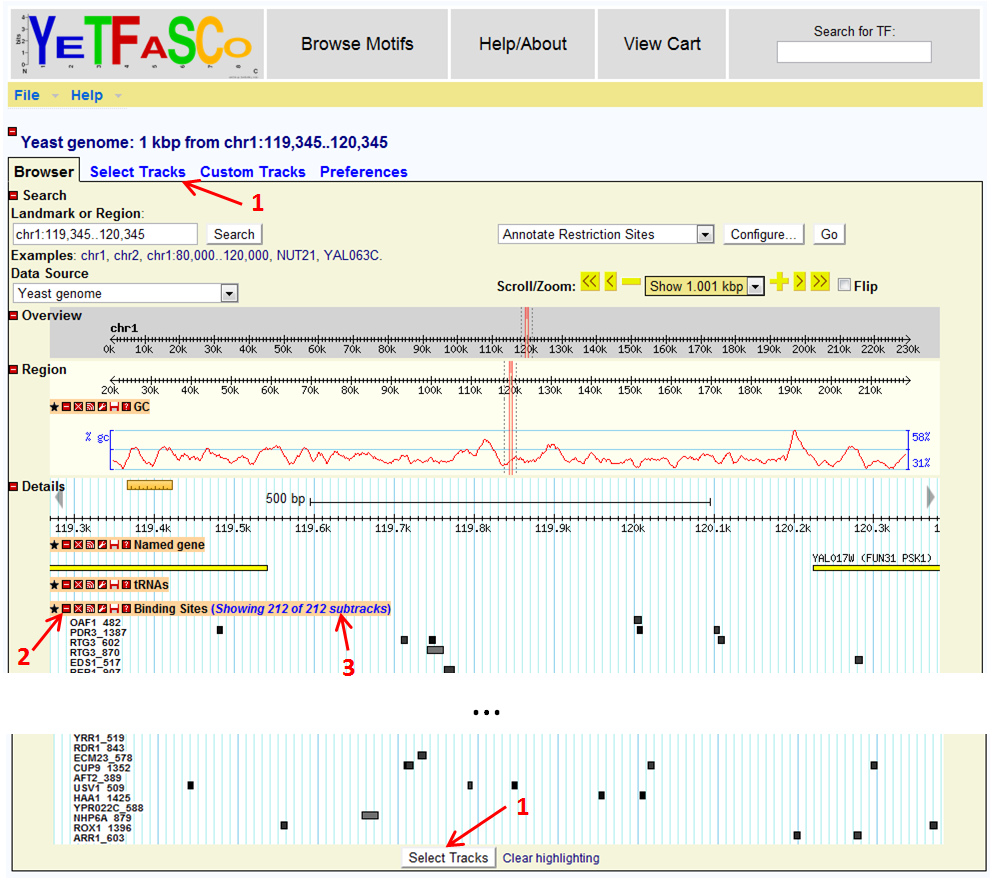
Here, you can select your tracks of interest (1), minimize, and perform other track-specific functions (2), and select subtracks (3). Subtracks are especially handy when you're only interested in a subset of transcription factors. Presently, the genome browser binding sites represent all binding sites that score at least 75% of the maximum possible PWM score. Also, if transcription factors are selected, but there are no binding sites in the browsed locality, the empty subtracks will be hidden.
You should now have a good idea how to navigate the site, but if you run into trouble, the Help section has a more detailed explanation of the site's functionality.